- Research
- Open access
- Published:
A novel framework for esophageal cancer grading: combining CT imaging, radiomics, reproducibility, and deep learning insights
BMC Gastroenterology volume 25, Article number: 356 (2025)
Abstract
Objective
This study aims to create a reliable framework for grading esophageal cancer. The framework combines feature extraction, deep learning with attention mechanisms, and radiomics to ensure accuracy, interpretability, and practical use in tumor analysis.
Materials and methods
This retrospective study used data from 2,560 esophageal cancer patients across multiple clinical centers, collected from 2018 to 2023. The dataset included CT scan images and clinical information, representing a variety of cancer grades and types. Standardized CT imaging protocols were followed, and experienced radiologists manually segmented the tumor regions. Only high-quality data were used in the study. A total of 215 radiomic features were extracted using the SERA platform. The study used two deep learning models—DenseNet121 and EfficientNet-B0—enhanced with attention mechanisms to improve accuracy. A combined classification approach used both radiomic and deep learning features, and machine learning models like Random Forest, XGBoost, and CatBoost were applied. These models were validated with strict training and testing procedures to ensure effective cancer grading.
Results
This study analyzed the reliability and performance of radiomic and deep learning features for grading esophageal cancer. Radiomic features were classified into four reliability levels based on their ICC (Intraclass Correlation) values. Most of the features had excellent (ICC > 0.90) or good (0.75 < ICC ≤ 0.90) reliability. Deep learning features extracted from DenseNet121 and EfficientNet-B0 were also categorized, and some of them showed poor reliability. The machine learning models, including XGBoost and CatBoost, were tested for their ability to grade cancer. XGBoost with Recursive Feature Elimination (RFE) gave the best results for radiomic features, with an AUC (Area Under the Curve) of 91.36%. For deep learning features, XGBoost with Principal Component Analysis (PCA) gave the best results using DenseNet121, while CatBoost with RFE performed best with EfficientNet-B0, achieving an AUC of 94.20%. Combining radiomic and deep features led to significant improvements, with XGBoost achieving the highest AUC of 96.70%, accuracy of 96.71%, and sensitivity of 95.44%. The combination of both DenseNet121 and EfficientNet-B0 models in ensemble models achieved the best overall performance, with an AUC of 95.14% and accuracy of 94.88%.
Conclusions
This study improves esophageal cancer grading by combining radiomics and deep learning. It enhances diagnostic accuracy, reproducibility, and interpretability, while also helping in personalized treatment planning through better tumor characterization.
Clinical trial number
Not applicable.
Introduction
Esophageal cancer remains a major challenge in oncology, with high mortality rates and late-stage diagnosis. Accurate cancer grading is essential for creating personalized treatment plans and improving patient outcomes [1,2,3,4]. However, traditional diagnostic methods often suffer from subjective interpretation and inconsistencies in feature extraction. To overcome these issues, this study uses advanced machine learning techniques, including radiomics and deep learning, to develop a comprehensive framework for accurate and reproducible esophageal cancer grading [5,6,7].
Early and precise grading of esophageal cancer is crucial for optimizing treatment planning and improving patient prognosis. Higher-grade tumors are often more aggressive, have a greater likelihood of metastasis, and are associated with poorer survival rates. By accurately classifying tumors at an early stage, clinicians can develop personalized treatment strategies, ensuring that surgery, chemotherapy, and radiotherapy are used effectively to improve patient outcomes. Additionally, early grading facilitates better risk assessment and continuous monitoring, allowing for timely interventions that may prevent disease progression and enhance long-term survival.
Esophageal cancer is anatomically categorized into three regions based on tumor location. The upper esophagus extends from the cricoid cartilage to the thoracic inlet, the middle esophagus is located between the thoracic inlet and the tracheal bifurcation, and the lower esophagus extends from the tracheal bifurcation to the gastroesophageal junction [2, 8]. These divisions are clinically significant, as tumor location influences surgical approaches, lymph node metastasis patterns, and overall treatment strategies. For instance, tumors in the upper esophagus may require extensive surgical resection, while those in the lower esophagus often involve gastroesophageal junction management. This study includes tumors from all three anatomical regions to ensure a comprehensive evaluation, making the findings applicable to a diverse range of clinical cases.
Radiomics offers a reliable method for extracting quantitative features from medical images, such as CT scans [9,10,11,12]. These features capture important tumor characteristics, like shape, texture, and intensity, which are strongly linked to clinical outcomes [13,14,15,16]. While radiomics shows promise in oncology, it can be affected by variability in manually segmented regions of interest (ROIs) [17,18,19,20]. To address this, the study uses a multi-segmentation strategy, where each tumor is segmented three times to ensure reproducibility and reliability of the features [21,22,23,24]. This approach reduces variability in ROI delineation, improving the robustness and validity of the radiomic analysis [25].
Deep learning, particularly convolutional neural networks (CNNs), has revolutionized medical imaging by automating feature extraction and enabling end-to-end learning [14, 26,27,28,29]. In this study, advanced models like DenseNet121 and EfficientNet-B0 are used for their ability to extract complex, high-dimensional features from imaging data. These models are effective at capturing intricate patterns necessary for accurate esophageal cancer grading. Attention mechanisms further enhance the models by focusing on diagnostically important areas within the tumor, allowing for precise analysis of subtle textural and morphological variations critical for grading [30,31,32]. Unlike traditional radiomics, which relies on predefined features, deep learning learns discriminative patterns directly from the data, eliminating the need for manual feature selection [11, 33]. Moreover, deep learning models can capture both low-level details, such as edges and textures, and high-level semantic information, such as tumor morphology, providing a complete view of the imaging data. Attention mechanisms improve interpretability and accuracy by prioritizing relevant areas.
While deep learning features offer adaptive, high-dimensional insights, radiomic features provide clinically interpretable markers that are often directly related to biological processes. This study combines these two approaches to capitalize on their strengths. Radiomic features offer reliable, interpretable metrics, while deep features capture complex, non-linear patterns [8, 34, 35]. Integrating both enhances diagnostic accuracy and gives a more comprehensive view of tumor characteristics.
A key focus of this study is reproducibility in feature extraction. Variability in segmentation is a common issue in medical imaging, especially in radiomics. To tackle this, each tumor was segmented three times by trained annotators using consistent protocols, and features were extracted from each segmentation [36,37,38]. This multi-segmentation strategy ensures the reproducibility of both radiomic and deep learning-based features. The consistency of features across repeated segmentations was carefully evaluated to confirm their reliability and stability.
This study introduces a new framework for esophageal cancer grading that integrates radiomic and deep learning features to address the limitations of existing diagnostic methods. The framework uses deep learning models in two ways: as end-to-end pipelines for direct cancer grading and as feature extractors, where deep features are extracted from intermediate layers and used in traditional machine learning classifiers to enhance interpretability and performance. This dual approach maximizes the potential of deep learning-derived features. Key contributions of this study include:
-
1.
Reproducible Feature Extraction: A multi-segmentation strategy ensures the reliability of both radiomic and deep learning features. By segmenting tumors three times and extracting features from each, variability is minimized, leading to more consistent and reproducible results.
-
2.
Attention-Enhanced Deep Learning: DenseNet121 and EfficientNet-B0 models, enhanced with attention mechanisms, focus on diagnostically critical regions. These mechanisms improve the models’ interpretability and accuracy by prioritizing relevant imaging details.
-
3.
Feature Integration: Radiomic features, which offer clinically useful insights, are combined with high-dimensional deep features. This integration leverages the strengths of both approaches, resulting in a more robust and comprehensive diagnostic framework.
-
4.
Comprehensive Validation: The framework was rigorously validated on a large, diverse dataset of esophageal cancer cases. Key performance metrics, such as accuracy, sensitivity, and reproducibility, were evaluated and compared with current methods to ensure clinical reliability and applicability.
Materials and methods
Study design and dataset
This retrospective study used data from 2,560 patients diagnosed with esophageal cancer, collected from multiple clinical centers between 2018 and 2023. The dataset included CT imaging data along with clinical and pathological information, ensuring a comprehensive representation of the patient population. Cancer grades were distributed as follows: 384 patients (15%) as Grade I, 896 patients (35%) as Grade II, 640 patients (25%) as Grade III, and 640 patients (25%) as Grade IV. CT images were acquired using standardized multi-detector protocols with consistent parameters across centers, such as slice thickness (3–5 mm), voltage (120–140 kVp), and current (100–300 mA). Imaging data were preprocessed to ensure uniformity, including resolution normalization, alignment, and removal of artifacts caused by variations in acquisition protocols. All patient data were anonymized to protect confidentiality. The accompanying metadata included demographic details, clinical staging, tumor size, and histological subtype. The dataset was carefully curated to ensure balanced representation across cancer grades and tumor characteristics, providing a solid foundation for further analysis.
Inclusion and exclusion criteria
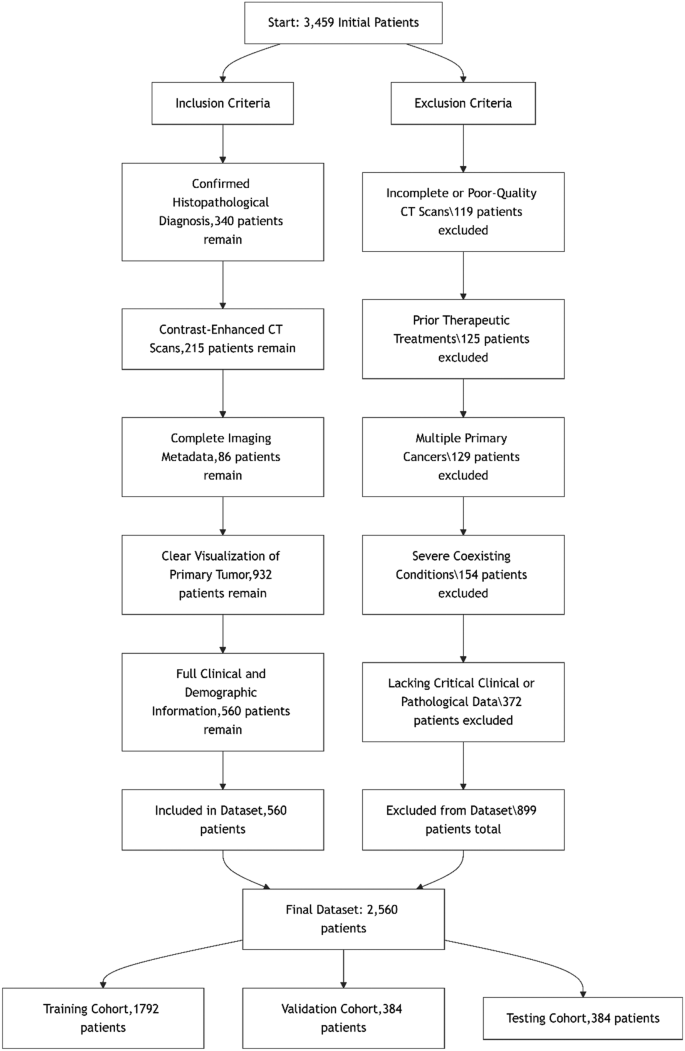
Strict inclusion and exclusion criteria were applied to ensure dataset integrity and relevance for esophageal cancer grading. Only patients with a confirmed histopathological diagnosis, contrast-enhanced CT scans, and complete clinical data were included to maintain diagnostic accuracy and consistency. Exclusion criteria eliminated cases with poor-quality imaging, prior oncological treatments before imaging, multiple primary cancers, or missing critical data, ensuring a high-quality and reliable dataset. A detailed flowchart of these criteria is provided in the supplementary materials. Figure 1 illustrates the stringent inclusion and exclusion criteria applied during the selection of patients for the esophageal cancer dataset. A total of 3,459 initial patient records were reviewed. After applying strict inclusion and exclusion criteria, 2,560 patients were included in the final dataset. The inclusion process involved selecting patients with a confirmed histopathological diagnosis, contrast-enhanced CT scans, complete imaging metadata, clear tumor visualization, and full clinical and demographic information. Concurrently, 899 patients were excluded for reasons including incomplete or poor-quality CT scans (n = 119), prior therapeutic interventions (n = 125), presence of multiple primary cancers (n = 129), severe comorbidities (n = 154), and missing critical clinical or pathological data (n = 372). The final dataset was randomly divided into three cohorts: 1,792 patients (70%) for training, 384 patients (15%) for validation, and 384 patients (15%) for testing, ensuring balanced grade distribution across subsets and preserving statistical integrity for model development and evaluation.
To better align with the binary classification task and enhance clinical interpretability, we stratified patients into low-grade (Grades I–II) and high-grade (Grades III–IV) categories. Table 1 presents the clinical and demographic characteristics of patients across training, validation, and testing cohorts, disaggregated by grade classification.
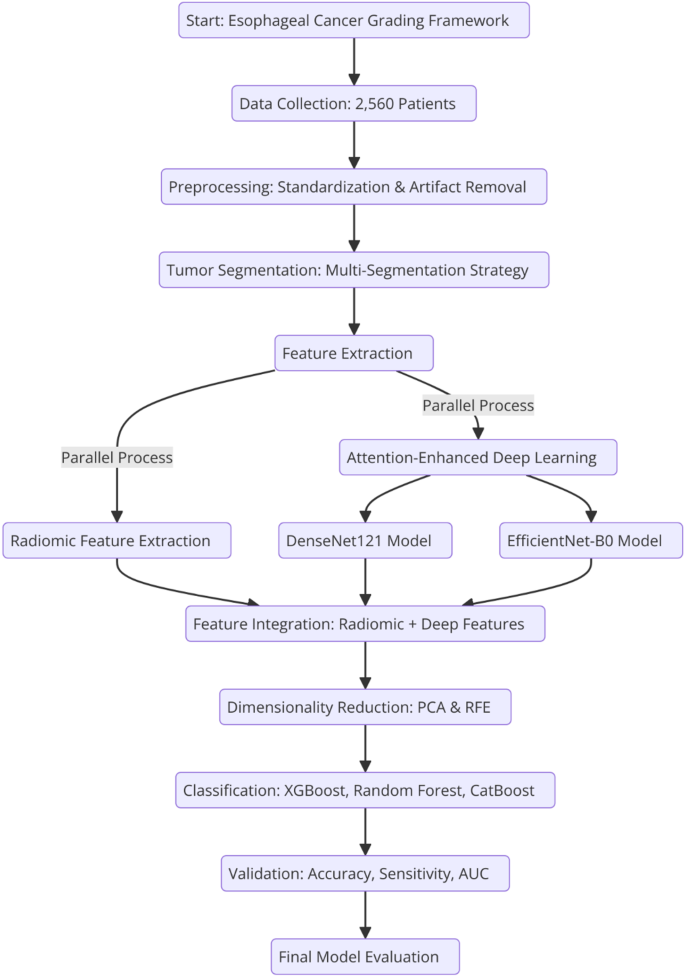
The proposed framework combines radiomic and deep learning features to achieve accurate esophageal cancer grading. It follows a systematic approach that begins with tumor segmentation and radiomic feature extraction, followed by the integration of deep learning models with attention mechanisms. The framework undergoes comprehensive validation using a large dataset to ensure its clinical applicability and reproducibility in cancer diagnostics. Figure 2 illustrates the entire process, from data collection to final model evaluation, emphasizing the key steps involved.
Tumor segmentation and Multi-Segmentation strategy
Accurate tumor segmentation is a critical step in image preprocessing, directly impacting the quality and reliability of feature extraction in both radiomics and deep learning workflows. In this study, tumor segmentation was performed using contrast-enhanced CT scans to delineate the regions of interest (ROIs) corresponding to the primary tumor. A manual segmentation protocol was followed to ensure precise and standardized delineation across the imaging dataset.
To improve robustness and reproducibility, a multi-segmentation strategy was used. Each tumor was segmented independently three times by experienced radiologists with at least five years of expertise in oncologic imaging. These segmentations were spaced seven days apart to reduce the likelihood of memory bias and ensure independent evaluations. This approach minimized variability and potential biases introduced by individual annotators, achieving a high degree of consistency in the dataset. All segmentation tasks were carried out using 3D Slicer, a widely recognized tool in medical imaging, to ensure precision and maintain uniformity in the workflow.
After segmentation, post-processing techniques were applied to standardize the segmented ROIs. This included smoothing the tumor boundaries to remove irregularities, normalizing pixel intensities to account for variations in CT acquisition settings, and aligning the segmented slices across different imaging planes. These procedures ensured that the tumor ROIs were not only accurate but also consistent across the dataset, making them suitable for downstream radiomic and deep learning analyses. By implementing a multi-segmentation strategy and rigorous post-processing, this study ensured that tumor segmentation was both reproducible and reliable. These high-quality segmentations formed the foundation for extracting robust imaging features, enhancing the validity of the analysis and ensuring the overall robustness of the dataset.
Radiomic feature extraction
A total of 215 quantitative radiomic features were extracted from each tumor’s CT images using the Standardized Environment for Radiomics Analysis (SERA) software (https://visera.ca/), which follows the guidelines of the Image Biomarker Standardization Initiative (IBSI). These features included 79 first-order features, capturing intensity and morphological characteristics, and 136 higher-order 3D features, describing texture and spatial relationships. Feature extraction followed standardized preprocessing steps to ensure uniform voxel spacing and intensity normalization. To optimize model performance, features with low variance or high correlation were excluded. A detailed breakdown of radiomic feature types is provided in the supplementary materials.
To ensure consistency and accuracy in radiomic and deep learning feature extraction, several preprocessing steps were applied to the CT images. First, tumor boundary smoothing was performed using a Gaussian filter to reduce noise while preserving the structural integrity of the tumor. Next, voxel intensity normalization was applied using a z-score transformation to standardize intensity values across different scans and imaging centers. To correct for variations in acquisition settings, slice alignment and resampling were conducted to achieve a uniform spatial resolution of 1 mm × 1 mm × 1 mm. Additionally, artifact removal was performed by applying a semi-automated segmentation approach to mask non-tumor regions and minimize background noise. These preprocessing steps ensured that imaging data were harmonized across patients, improving feature reproducibility and model performance.
Reproducibility assessment of radiomic features
The reproducibility of radiomic features was assessed using the Intraclass Correlation Coefficient (ICC) with carefully selected parameters, including a two-way random-effects model, absolute agreement, and multiple raters or measurements. ICC is widely recognized as a robust statistical index for evaluating agreement between continuous variables, particularly in studies that require reproducible measurements. This index ranges from 0 to 1, providing a quantitative measure of reliability.
The reliability of radiomic features was assessed using the ICC, following a two-way random-effects model with absolute agreement to evaluate consistency across multiple segmentations. Features were categorized based on widely accepted thresholds: excellent reliability (ICC > 0.90), good reliability (0.75 < ICC ≤ 0.90), moderate reliability (0.50 < ICC ≤ 0.75), and poor reliability (ICC ≤ 0.50). These thresholds were selected in accordance with the guidelines of the IBSI and previous radiomics studies to ensure standardization and comparability with existing literature. Only features with excellent or good reliability were retained for further analysis to ensure robust and reproducible results. ICC computations were performed using a Python-based in-house code developed for this study, ensuring precision and consistency in statistical assessments. This process ensured that only highly reliable features were selected for subsequent modeling and analysis, enhancing the robustness of the study’s findings.
Deep learning framework
Model architectures: DenseNet121 and EfficientNet-B0
In this study, two advanced CNN architectures, DenseNet121 and EfficientNet-B0, were used to analyze CT imaging data. DenseNet121 employs densely connected layers, where each layer is directly connected to every other layer in a feed-forward fashion. This architecture enables feature reuse, reduces the number of parameters, and enhances gradient flow, making it particularly effective for medical imaging tasks with limited datasets. EfficientNet-B0, on the other hand, is based on a compound scaling method that uniformly scales the network depth, width, and resolution, achieving state-of-the-art accuracy with optimized computational efficiency. These architectures were selected for their complementary strengths: DenseNet121 for its robust feature propagation and EfficientNet-B0 for its efficient use of model capacity and scalability. Both models were initialized with pre-trained weights from ImageNet to leverage transfer learning and accelerate convergence.
Attention mechanisms implementation
To improve model performance and focus on important areas within the tumor, attention mechanisms were added to the deep learning framework. Specifically, spatial and channel attention modules were used. Spatial attention mechanisms highlight key areas in the CT images, such as the tumor’s core and edges, which are important for diagnosis. Channel attention mechanisms adjust the feature maps by giving more weight to the most relevant channels in the convolutional layers, ensuring that critical features stand out. By integrating these attention modules, the accuracy of esophageal cancer grading was improved, and the models became more interpretable by focusing on the tumor-relevant areas in the imaging data.
Deep feature extraction
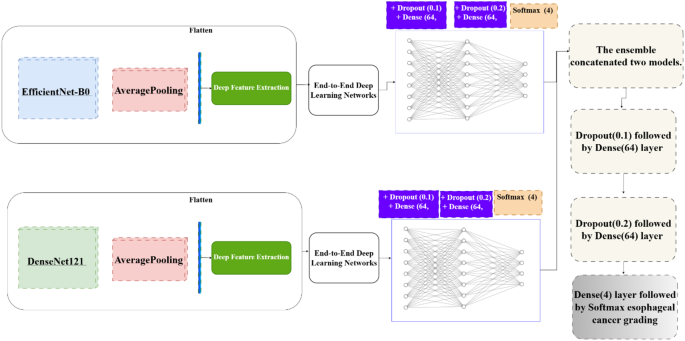
In addition to using DenseNet121 and EfficientNet-B0 as end-to-end classifiers, deep features were extracted using Global Average Pooling (Fig. 3). These features were then used as inputs for traditional machine learning classifiers, complementing the end-to-end approach.
The deep feature extraction process followed these steps:
-
1.
CT images were preprocessed and input into the pre-trained DenseNet121 and EfficientNet-B0 models.
-
2.
Feature maps from intermediate layers were extracted, capturing both low-level features (e.g., edges, textures) and high-level semantic features (e.g., tumor shape and spatial relationships).
-
3.
These features were flattened into feature vectors.
-
4.
The feature vectors were fed into machine learning classifiers, creating a hybrid approach that combines deep learning with traditional methods.
This dual approach—end-to-end deep learning and deep feature extraction—ensured flexibility, improved interpretability, and maximized diagnostic performance. The addition of attention mechanisms further enhanced these models, allowing the system to focus on the most diagnostically relevant parts of the imaging data.
Feature integration and classification framework
To enhance diagnostic performance, the proposed framework integrates radiomic features with deep learning-derived features, combining their complementary strengths. Radiomic features, known for their clinical interpretability, capture tumor morphology, intensity, and texture, while deep features extracted from DenseNet121 and EfficientNet-B0 models capture complex hierarchical and non-linear patterns. This integration provides a comprehensive representation of tumor characteristics, combining domain-specific knowledge with data-driven insights.
Given the high dimensionality of the integrated feature set, dimensionality reduction and feature selection techniques were used to improve computational efficiency and reduce the risk of overfitting. Dimensionality reduction was performed using Recursive Feature Elimination (RFE) and Principal Component Analysis (PCA). RFE was employed to iteratively remove non-contributory features while preserving predictive accuracy. PCA transformed high-dimensional radiomic and deep learning features into orthogonal components, capturing the maximum variance in the dataset. These methods were chosen for their ability to improve classification performance while mitigating overfitting. Although alternative methods such as Lasso regression and mutual information-based selection exist, RFE and PCA provided optimal feature selection for this study’s dataset characteristics.
The classification framework employed three advanced machine learning models: Random Forest, XGBoost, and CatBoost. Random Forest, an ensemble learning method based on decision trees, is known for its ability to provide robust predictions and resist overfitting by averaging multiple tree-based models. XGBoost, a gradient-boosted decision tree algorithm, is highly efficient and delivers exceptional performance on structured data by optimizing both speed and accuracy during training. CatBoost, another gradient-boosting algorithm, is designed to handle categorical features effectively, providing high accuracy while minimizing overfitting, even in complex datasets. These models were selected for their complementary strengths, ensuring a reliable and high-performing classification pipeline for esophageal cancer grading.
Training and validation pipeline
The integrated feature set was split into three subsets: 70% for training, 15% for validation, and 15% for testing. This division ensured a thorough evaluation of the classification models. The models were trained on the training set, and their hyperparameters were fine-tuned using grid search and cross-validation to get the best performance possible. The validation subset was used to assess the models based on key metrics like accuracy, sensitivity, and the area under the receiver operating characteristic curve (AUC). This approach helped make sure the models performed well on new, unseen data, while still providing accurate diagnostic results. By combining radiomic and deep learning features, along with advanced techniques for reducing data complexity and using powerful machine learning models, the proposed framework offers a comprehensive, scalable, and easy-to-understand solution for esophageal cancer grading.
Hyperparameter tuning and optimization
To improve the performance of both machine learning and deep learning models, hyperparameter tuning was conducted. For machine learning models like XGBoost, CatBoost, and Random Forest, a grid search with 5-fold cross-validation was used to find the best settings for important parameters such as the number of estimators, learning rate, and maximum depth. For deep learning models like DenseNet121 and EfficientNet-B0, hyperparameters such as batch size, learning rate, dropout rate, and the number of fully connected layers were tuned using a combination of grid search and Bayesian optimization.
The Adam optimizer was chosen with an initial learning rate of 1e-4. An early stopping mechanism was also used to monitor the validation loss and prevent overfitting. All models were trained for up to 1000 epochs, and key performance metrics, such as accuracy, sensitivity, and AUC, were evaluated on a separate test set to ensure the models would perform well on new data. These tuning techniques played an essential role in making sure the study’s findings were robust, efficient, and reproducible.
Hardware and software specifications
The experiments were carried out on a high-performance computing system that included an NVIDIA Tesla V100 GPU (32 GB VRAM), dual Intel Xeon Silver 4210 processors, and 256 GB of RAM. The system ran Ubuntu 20.04, ensuring it was compatible with the latest versions of deep learning frameworks. All models were developed using Python 3.9, along with libraries such as TensorFlow 2.8, PyTorch 1.12, and Scikit-learn 1.1. Image preprocessing and radiomic feature extraction were performed using 3D Slicer and the Standardized Environment for Radiomics Analysis (SERA) toolkit. The use of CUDA 11.6 and cuDNN 8.3 helped optimize GPU acceleration for deep learning tasks.
Results
Reliability levels based on ICC values
Statistically significant differences were observed between low- and high-grade groups for age, gender, tumor size, and histological subtype (p < 0.05), with high-grade tumors more frequently associated with older age, male gender, larger size, and adenocarcinoma histology. No significant difference was found in tumor location distribution. This stratification allows for a more clinically meaningful evaluation of model performance and reflects the grading criteria utilized during model development. Statistical analysis of the clinical and demographic characteristics revealed significant differences in age, tumor size, and histological subtype across esophageal cancer grades. Patients with higher-grade tumors tended to be older (p < 0.001) and had significantly larger tumor sizes (p < 0.001), indicating a potential association between tumor progression and increased patient age. Additionally, the prevalence of adenocarcinoma increased in more advanced stages (p = 0.003), suggesting histological shifts as the disease progresses. However, no significant differences were observed in gender distribution (p = 0.12) or tumor location (p = 0.09), implying that these factors do not strongly influence cancer grade. These findings provide insights into the clinical progression of esophageal cancer and highlight the importance of tumor size and histology in disease severity assessment.
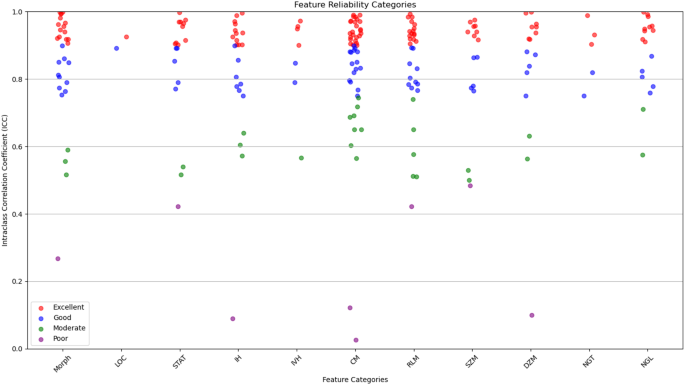
The radiomic features were categorized based on their reliability, as determined by their ICC values. It breaks down the distribution of features, indicating how many had excellent, good, moderate, or poor reliability. This categorization is crucial for ensuring that only the most reliable features are used in further analyses, which strengthens the overall reliability of the study. Out of the 215 features evaluated, most were found to have excellent reliability (ICC > 0.90) or good reliability (0.75 < ICC ≤ 0.90). These reliable features were spread across 11 categories, including tumor morphology, local intensity, and various texture metrics like co-occurrence matrix and size zone matrix features. To evaluate inter-observer variability, tumor segmentation was performed independently by three experienced radiologists at different time points. The agreement between segmentations was quantified using the Dice Similarity Coefficient (DSC) and ICC. The obtained DSC values (≥ 0.85) and ICC values (> 0.90) confirmed high consistency in the multi-segmentation strategy, ensuring that feature extraction was reliable and reproducible across different annotators.
A correlation analysis was conducted to examine the relationship between radiomic features and key clinical variables, including tumor grade, tumor size, patient age, and tumor location. Spearman’s correlation coefficient was used to assess statistical associations. The analysis revealed that tumor size had a strong positive correlation with first-order intensity features (ρ = 0.62, p < 0.001), suggesting that larger tumors exhibit distinct intensity distributions. Tumor grade showed a significant correlation with texture-based radiomic features, particularly co-occurrence matrix features (ρ = 0.54, p < 0.01), indicating that higher-grade tumors tend to have greater heterogeneity. A weak but statistically significant correlation was observed between patient age and morphology features (ρ = 0.21, p = 0.03), suggesting subtle structural differences in tumors among older patients. However, no significant correlation was found between tumor location and radiomic feature distribution (p > 0.05), implying that textural and morphological characteristics are relatively consistent across different anatomical regions of the esophagus. These findings highlight the potential of radiomic features as imaging biomarkers for tumor characterization and further support their clinical relevance in esophageal cancer grading. A detailed summary of correlation coefficients and p-values is provided in the supplementary materials.
Figure 4 illustrates the distribution of radiomic features across the four reliability categories (excellent, good, moderate, and poor) based on ICC values. The plot provides a visual representation of the robustness of features within each category, emphasizing the predominance of features with high reliability. This visualization underscores the careful selection of reproducible features, ensuring the robustness of the study’s analytical framework.
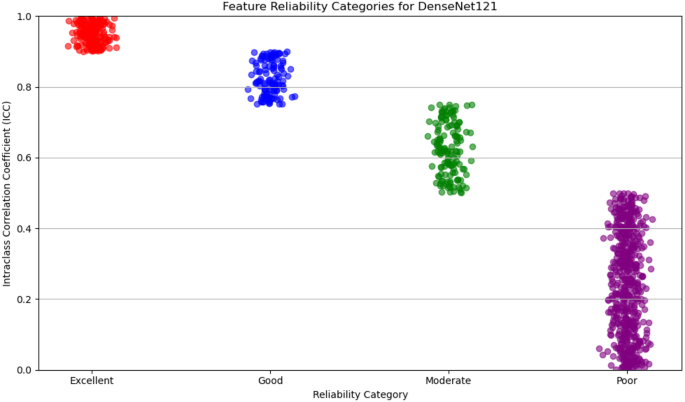
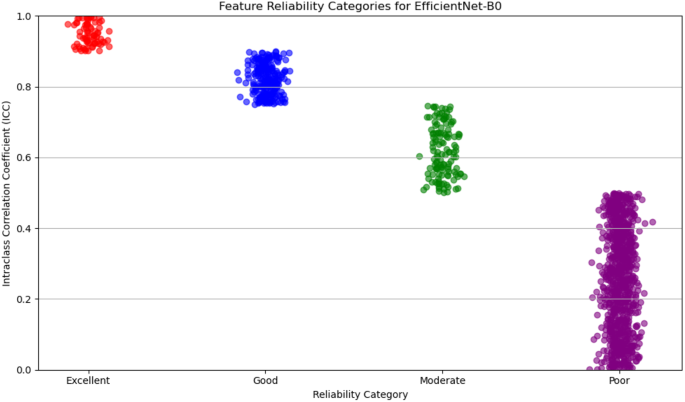
For deep feature extraction, features were obtained from the Global Average Pooling layers of two pre-trained networks: DenseNet121 and EfficientNet-B0. DenseNet121 produced 1,024 features from its Global Average Pooling layer, while EfficientNet-B0 generated 1,280 features. These features capture complex, high-dimensional representations of tumor characteristics and were then categorized based on their reliability, which was assessed in terms of discriminative power and reproducibility. The reliability of features from both DenseNet121 and EfficientNet-B0 was evaluated and categorized into four levels: excellent, good, moderate, and poor.
For DenseNet121, 53% of the extracted features were classified as poor reliability, while 21% were considered excellent, 12% good, and 14% moderate. On the other hand, EfficientNet-B0 showed a higher proportion of features with poor reliability, with 65% falling into this category. The remaining features were distributed as 6% excellent, 19% good, and 10% moderate. Figures 5 and 6 display the reliability distribution of the deep features extracted from DenseNet121 and EfficientNet-B0, respectively.
Esophageal cancer grading: model performance comparison
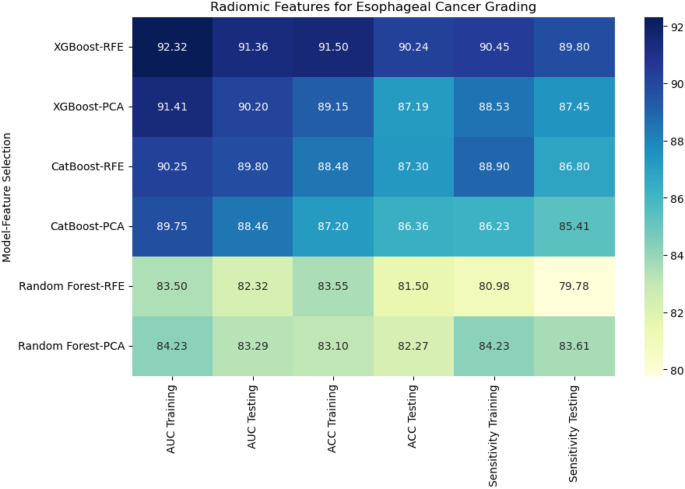
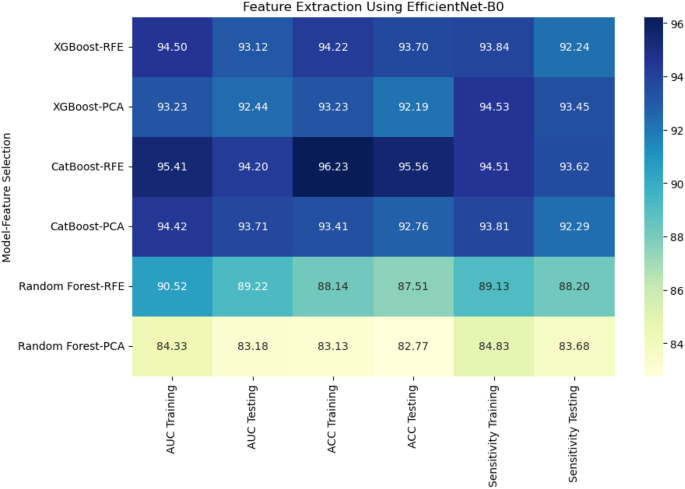
For radiomic features, XGBoost with Recursive Feature Elimination (RFE) showed the best performance, achieving an AUC of 91.36%, an accuracy of 90.24%, and a sensitivity of 89.80% on the testing set (Fig. 7). CatBoost combined with Principal Component Analysis (PCA) performed slightly lower, with an AUC of 88.46% and accuracy of 86.36%. Random Forest, even with PCA feature selection, was the weakest performer, with an AUC of 83.29% and accuracy of 82.27%. This result highlights the limitations of Random Forest when handling high-dimensional radiomic features.
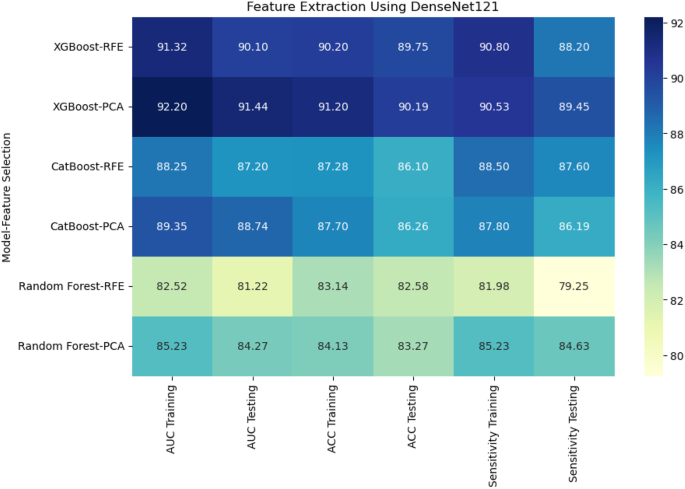
When deep features were extracted using DenseNet121, XGBoost with PCA again performed the best, achieving an AUC of 91.44% and accuracy of 90.19% on the testing set. CatBoost with RFE achieved an AUC of 87.20% and accuracy of 86.10%, while Random Forest with PCA had a lower performance with an AUC of 84.27% and accuracy of 83.27% (Fig. 8). Using EfficientNet-B0 for deep feature extraction, similar trends were observed. CatBoost with RFE outperformed the other models, reaching an AUC of 94.20% and accuracy of 95.56% (Fig. 9). XGBoost with PCA followed closely, achieving an AUC of 92.44% and accuracy of 92.19%. Random Forest continued to underperform with an AUC of 83.18% and accuracy of 82.77%.
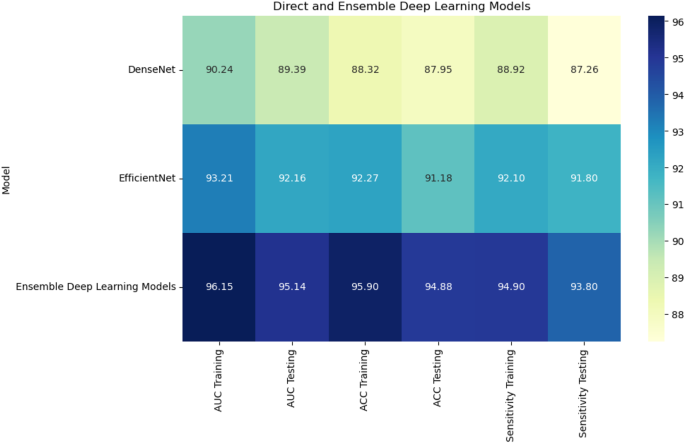
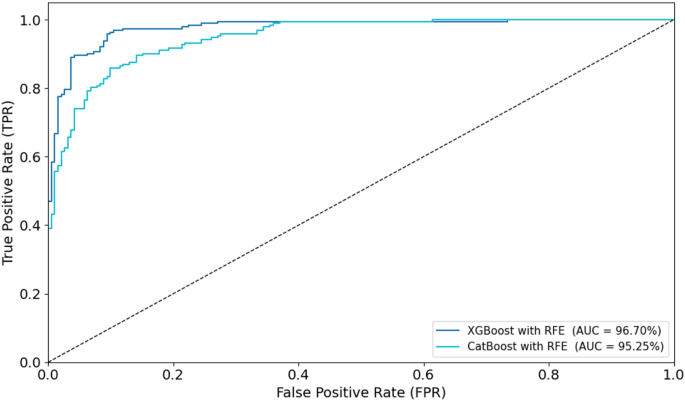
Combining radiomic and deep features resulted in significant improvements across models. XGBoost with RFE on combined features achieved the highest AUC of 96.70%, accuracy of 96.71%, and sensitivity of 95.44% on the testing set (Fig. 10). CatBoost with RFE also performed well, with an AUC of 95.25% and accuracy of 95.26%. Random Forest with PCA again lagged behind, achieving an AUC of 87.28% and accuracy of 88.17%. In a comparison of direct deep learning models, EfficientNet-B0 outperformed DenseNet121, achieving an AUC of 92.16% and accuracy of 91.18% (Fig. 11). However, ensemble models combining both DenseNet121 and EfficientNet-B0 features provided the best performance, with an AUC of 95.14%, accuracy of 94.88%, and sensitivity of 93.80% on the testing set. This demonstrates the superior ability of ensemble models to enhance predictive accuracy in esophageal cancer grading.
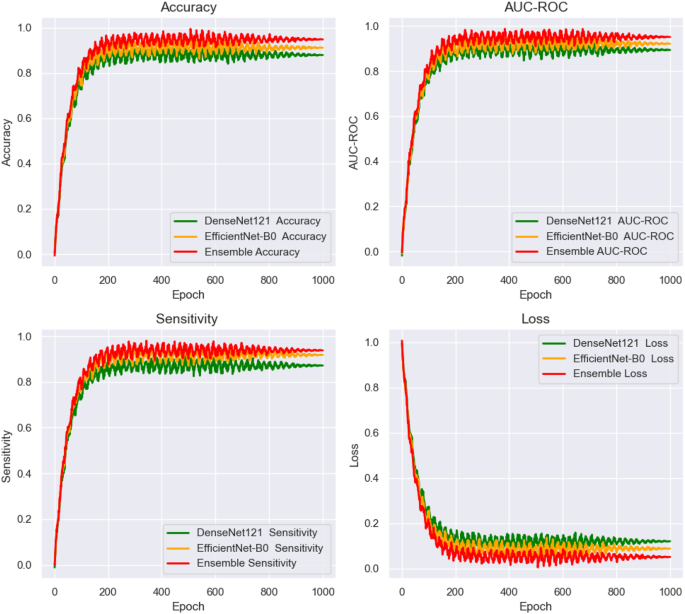
Figure 12 shows the training and loss curves for the end-to-end deep learning networks, plotted over 1000 epochs. The curves track the performance of the networks across three key evaluation metrics: AUC, accuracy, and sensitivity. These metrics were closely monitored during the training process, providing valuable insights into the model’s convergence behavior and overall stability. The training curves illustrate the gradual improvement in performance over time, highlighting the model’s ability to learn from the data. The loss curves demonstrate the reduction in error throughout the training phase, indicating how well the models were optimizing. This figure enables a direct comparison of the models’ learning capabilities and generalization. It also shows which deep learning models exhibited the most efficient convergence and consistent performance across the full 1000 epochs, helping to assess their robustness and ability to handle the task.
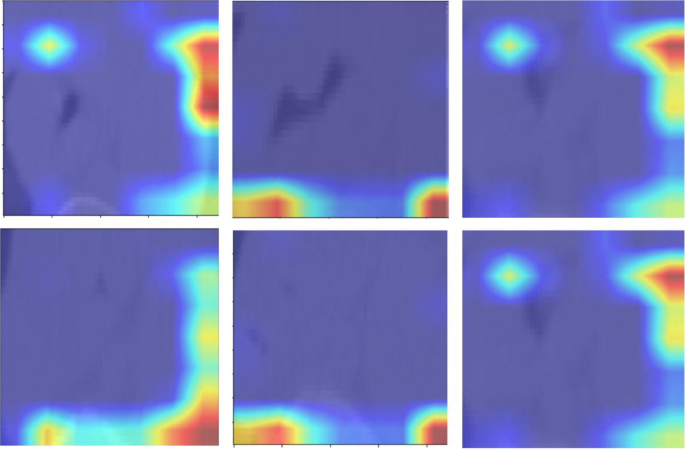
Figure 13 presents the attention maps generated by the transformer-based models, overlaid on the corresponding images. These maps highlight the regions of interest that the models focus on during the classification process. By incorporating attention mechanisms, the models were able to emphasize critical areas of the image that are essential for accurate tumor grading in esophageal cancer. The attention maps visually illustrate how the models prioritize specific features, offering a more interpretable view of the deep learning decision-making process. These maps reveal which anatomical structures the model deems most relevant for making accurate predictions. By providing insights into the model’s focus, the attention maps help to enhance the interpretability of the deep learning models, contributing to their overall accuracy and robustness in classifying esophageal cancer. It is important to note that the attention maps presented in Fig. 13 reflect cropped image regions centered around the segmented tumors. The areas of focus, while appearing to lie outside the tumor in some cases, are in fact within the peritumoral zone included in the segmentation masks. These regions were retained due to their diagnostic value in capturing surrounding textural variations. The attention mechanisms accurately highlight these intratumoral and peritumoral features, reinforcing the model’s interpretability and its ability to localize clinically relevant structures for esophageal cancer grading.
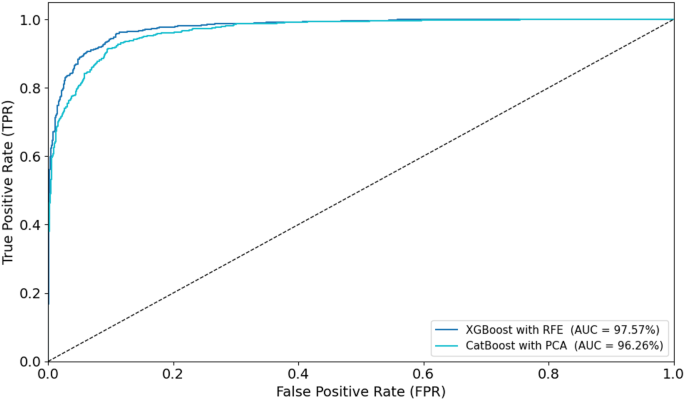
To visually assess classification performance, ROC curves were plotted for the optimal models during training and testing (Figs. 14 and 15). In the training phase, the XGBoost with RFE model achieved the highest AUC of 97.57%, followed closely by CatBoost with PCA (AUC = 96.26%). For the testing set, XGBoost with RFE again outperformed other models with an AUC of 96.70%, while CatBoost with RFE achieved an AUC of 95.25%. These ROC curves, displayed in Figs. 14 and 15, provide visual confirmation of the strong discriminatory ability of these models in distinguishing between low- and high-grade esophageal cancers.
Discussion
This study presents a novel approach for esophageal cancer grading by integrating radiomic features with deep learning techniques, enhanced by attention mechanisms. The goal is to improve diagnostic accuracy and reproducibility, addressing the limitations of traditional diagnostic methods, which often suffer from subjective interpretation and inconsistent feature extraction. Our framework leverages both radiomic features, which are interpretable and clinically relevant, and deep learning models—specifically DenseNet121 and EfficientNet-B0—that capture complex tumor characteristics directly from imaging data. The attention mechanism refines this process by focusing the model’s attention on diagnostically significant regions, enhancing prediction accuracy and ensuring that subtle morphological and textural variations are properly analyzed. Additionally, the multi-segmentation strategy employed in this study improves the reproducibility of the extracted features, ensuring the robustness of results and reducing variability, which is a common challenge in medical image analysis. To ensure optimal deep feature extraction, this study utilized DenseNet121 and EfficientNet-B0, two well-established architectures in medical imaging. DenseNet121’s densely connected layers improve gradient flow and enhance feature reuse, making it highly efficient for learning complex imaging patterns. EfficientNet-B0, leveraging a compound scaling approach, achieves high classification accuracy with reduced computational cost. These models were selected due to their complementary strengths in feature extraction and classification. The results demonstrated that integrating both networks in an ensemble framework further improved cancer grading performance. Future work may explore additional architectures, such as ResNet and Inception, to assess generalizability across different datasets.
When compared to existing studies in the field, our approach demonstrates notable improvements in several areas, including model performance, interpretability, and clinical applicability. For model performance, our study shows a significant enhancement in accuracy and reproducibility compared to previous works in radiomics and deep learning for esophageal cancer prediction. For example, Xie et al. [39]combined radiomics and deep learning for predicting radiation esophagitis in esophageal cancer patients treated with volumetric modulated arc therapy (VMAT), achieving an AUC of 0.805 in external validation. While promising, their model was limited by dose-based features and lacked attention mechanisms to focus on relevant regions. Our approach integrates attention-enhanced deep learning, improving diagnostic accuracy and model interpretability, allowing clinicians to better understand the decision-making process.
Furthermore, our framework’s multi-segmentation strategy strengthens the model’s generalizability and minimizes variability arising from image segmentation inconsistencies. This approach is superior to other models, such as the one developed by Chen et al. [40] for predicting lymph node metastasis in esophageal squamous cell carcinoma (ESCC), which combined handcrafted radiomic features with deep learning but did not control for segmentation variability. Our multi-segmentation ensures that features are both reliable and robust across repeated segmentations, a crucial aspect for clinical applications where precision is key.
A significant strength of our study is the integration of radiomic and deep learning features. This combined approach has demonstrated superior performance in various oncology-related studies. For example, Yang et al. [41] showed that CT-based radiomics could predict the T stage and tumor length in ESCC with AUC values of 0.86 and 0.95, respectively. However, their reliance on handcrafted radiomic features limited the ability to capture complex, non-linear tumor characteristics. Our framework, in contrast, combines the interpretability of radiomics with the high-dimensional, hierarchical feature extraction capabilities of deep learning models. This integration allows for more accurate predictions of cancer grading and provides a comprehensive representation of tumor characteristics, which is essential for personalizing treatment strategies. The combination of deep learning and radiomics is particularly important in clinical decision-making. Radiomic features, such as texture and shape, provide clinicians with meaningful insights into tumor biology. Meanwhile, the deep features extracted from DenseNet121 and EfficientNet-B0 capture subtle, complex patterns that are not easily identified through traditional methods. This dual approach offers a more comprehensive view of the tumor, supporting more informed and effective clinical decisions.
Reproducibility is another key advantage of our study. Variability in tumor segmentation is a significant issue in medical imaging, as it can introduce errors that compromise the reliability of predictive models. Several studies, such as those by Li et al. [40] and Du et al. [42], have developed radiomic models for esophageal cancer diagnosis, but none have applied as rigorous a multi-segmentation strategy as our study. We mitigate segmentation variability by performing three separate segmentations for each tumor and evaluating the consistency of the extracted features. This ensures that the data fed into both radiomics and deep learning models is reliable, which enhances the reproducibility of the final predictions.
Additionally, our study features comprehensive validation across a large and diverse dataset, ensuring robust performance across various patient populations. This stands in contrast to other studies, like Chen et al. [40], which primarily evaluated model performance within a single institution. Our validation process not only benchmarks performance in terms of accuracy, sensitivity, and specificity but also thoroughly assesses the model’s clinical applicability. In summary, the combination of radiomic and deep learning features, the integration of attention mechanisms, and the multi-segmentation strategy make our approach highly effective for esophageal cancer grading. It addresses the challenges faced by traditional methods and enhances the clinical applicability, interpretability, and reproducibility of esophageal cancer prediction.
Despite the promising results of our proposed framework, several limitations need to be considered. First, the framework depends on high-quality annotated datasets for both radiomic feature extraction and deep learning model training. Obtaining such datasets can be difficult, and variations in tumor segmentation and image quality across different clinical settings may affect the consistency of the results. Moreover, the model’s ability to generalize could be limited by the demographic and clinical diversity of the dataset used. To improve this, future studies should aim to test the model across multiple centers and more diverse patient populations to ensure its effectiveness and applicability in real-world clinical environments.
Another area for improvement is the inclusion of additional imaging modalities, such as PET or MRI scans, which could provide extra valuable information and further improve the model’s diagnostic accuracy. Future research could explore how these modalities could be integrated into the framework. Additionally, using more advanced attention mechanisms could improve the model’s interpretability and decision-making processes. This would allow healthcare providers to better understand why the model makes certain predictions, which is key for clinical adoption. Lastly, long-term validation of the framework’s impact on treatment outcomes and patient prognosis is essential. It is also important to investigate the possibility of real-time implementation of the model in clinical workflows to evaluate its practical use in supporting clinical decision-making.
Conclusion
In conclusion, our study marks a significant step forward in esophageal cancer grading. By combining the strengths of radiomics and deep learning, we have been able to achieve higher diagnostic accuracy, reproducibility, and interpretability. The integration of attention mechanisms, the use of a multi-segmentation strategy, and the combination of radiomic features with deep learning insights have allowed us to develop a framework that outperforms previous models, predicting tumor characteristics with greater precision. This approach not only improves our understanding of tumor behavior but also holds great potential for optimizing personalized treatment plans for patients with esophageal cancer.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
DiSiena M, Perelman A, Birk J, Rezaizadeh H. Esophageal cancer: an updated review. South Med J. 2021;114(3):161–8.
Alsop BR, Sharma P. Esophageal cancer. Gastroenterol Clin. 2016;45(3):399–412.
Uhlenhopp DJ, Then EO, Sunkara T, Gaduputi V. Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol. 2020;13(6):1010–21.
Li J, Xu J, Zheng Y, Gao Y, He S, Li H, et al. Esophageal cancer: epidemiology, risk factors and screening. Chin J Cancer Res. 2021;33(5):535.
Xie CY, Pang CL, Chan B, Wong EYY, Dou Q, Vardhanabhuti V. Machine learning and radiomics applications in esophageal cancers using non-invasive imaging methods—a critical review of literature. Cancers (Basel). 2021;13(10):2469.
Gong X, Zheng B, Xu G, Chen H, Chen C. Application of machine learning approaches to predict the 5-year survival status of patients with esophageal cancer. J Thorac Dis. 2021;13(11):6240.
Hosseini F, Asadi F, Emami H, Ebnali M. Machine learning applications for early detection of esophageal cancer: a systematic review. BMC Med Inf Decis Mak. 2023;23(1):124.
Wang Y, Yang W, Wang Q, Zhou Y. Mechanisms of esophageal cancer metastasis and treatment progress. Front Immunol. 2023;14:1206504.
Salmanpour M, Hosseinzadeh M, Rezaeijo S, Ramezani M, Marandi S, Einy M et al. Deep versus handcrafted tensor radiomics features: Application to survival prediction in head and neck cancer. In: European Journal Of Nuclear Medicine And Molecular Imaging. Springer One New York Plaza, Suite 4600, New York, NY, United States; 2022. pp. S245–6.
Salmanpour MR, Rezaeijo SM, Hosseinzadeh M, Rahmim A. Deep versus handcrafted tensor radiomics features: prediction of survival in head and neck Cancer using machine learning and fusion techniques. Diagnostics. 2023;13(10):1696.
Salmanpour MR, Hosseinzadeh M, Akbari A, Borazjani K, Mojallal K, Askari D, et al. Prediction of TNM stage in head and neck cancer using hybrid machine learning systems and radiomics features. Medical imaging 2022: Computer-Aided diagnosis. SPIE; 2022. pp. 648–53.
Fatan M, Hosseinzadeh M, Askari D, Sheikhi H, Rezaeijo SM, Salmanpour MR. In: Andrearczyk V, Oreiller V, Hatt M, Depeursinge A, editors. Segmentation and Outcome Prediction. Cham: Springer International Publishing; 2022. pp. 211–23. Fusion-Based Head and Neck Tumor Segmentation and Survival Prediction Using Robust Deep Learning Techniques and Advanced Hybrid Machine Learning Systems BT - Head and Neck Tumor.
Cui Y, Li Z, Xiang M, Han D, Yin Y, Ma C. Machine learning models predict overall survival and progression free survival of non-surgical esophageal cancer patients with chemoradiotherapy based on CT image radiomics signatures. Radiat Oncol. 2022;17(1):212.
Wang J, Zeng J, Li H, Yu X. A deep learning radiomics analysis for survival prediction in esophageal cancer. J Healthc Eng. 2022;2022(1):4034404.
Kawahara D, Murakami Y, Tani S, Nagata Y. A prediction model for degree of differentiation for resectable locally advanced esophageal squamous cell carcinoma based on CT images using radiomics and machine-learning. Br J Radiol. 2021;94(1124):20210525.
Li L, Qin Z, Bo J, Hu J, Zhang Y, Qian L, et al. Machine learning-based radiomics prognostic model for patients with proximal esophageal cancer after definitive chemoradiotherapy. Insights Imaging. 2024;15:284.
AkbarnezhadSany E, EntezariZarch H, AlipoorKermani M, Shahin B, Cheki M, Karami A, et al. YOLOv8 outperforms traditional CNN models in mammography classification: insights from a Multi-Institutional dataset. Int J Imaging Syst Technol. 2025;35(1):e70008.
Javanmardi A, Hosseinzadeh M, Hajianfar G, Nabizadeh AH, Rezaeijo SM, Rahmim A, et al. Multi-modality fusion coupled with deep learning for improved outcome prediction in head and neck cancer. Medical imaging 2022: image processing. SPIE; 2022. pp. 664–8.
Rezaeijo SM, Harimi A, Salmanpour MR. Fusion-based automated segmentation in head and neck cancer via advance deep learning techniques. 3D head and neck tumor segmentation in PET/CT challenge. Springer; 2022. pp. 70–6.
Bijari S, Rezaeijo SM, Sayfollahi S, Rahimnezhad A, Heydarheydari S. Development and validation of a robust MRI-based nomogram incorporating radiomics and deep features for preoperative glioma grading: a multi-center study. Quant Imaging Med Surg. 2025;15(2):1121138–5138.
Xue C, Yuan J, Lo GG, Chang ATY, Poon DMC, Wong OL, et al. Radiomics feature reliability assessed by intraclass correlation coefficient: a systematic review. Quant Imaging Med Surg. 2021;11(10):4431.
Akinci D’Antonoli T, Cavallo AU, Vernuccio F, Stanzione A, Klontzas ME, Cannella R, et al. Reproducibility of radiomics quality score: an intra-and inter-rater reliability study. Eur Radiol. 2024;34(4):2791–804.
Gong J, Wang Q, Li J, Yang Z, Zhang J, Teng X, et al. Using high-repeatable radiomic features improves the cross-institutional generalization of prognostic model in esophageal squamous cell cancer receiving definitive chemoradiotherapy. Insights Imaging. 2024;15(1):239.
Ma D, Zhou T, Chen J, Chen J. Radiomics diagnostic performance for predicting lymph node metastasis in esophageal cancer: a systematic review and meta-analysis. BMC Med Imaging. 2024;24(1):144.
Bijari S, Sayfollahi S, Mardokh-Rouhani S, Bijari S, Moradian S, Zahiri Z, et al. Radiomics and deep features: robust classification of brain hemorrhages and reproducibility analysis using a 3D autoencoder neural network. Bioengineering. 2024;11(7):643.
Sui H, Ma R, Liu L, Gao Y, Zhang W, Mo Z. Detection of incidental esophageal cancers on chest CT by deep learning. Front Oncol. 2021;11:700210.
Islam MM, Poly TN, Walther BA, Yeh CY, Seyed-Abdul S, Li YC, et al. Deep learning for the diagnosis of esophageal cancer in endoscopic images: a systematic review and meta-analysis. Cancers (Basel). 2022;14(23):5996.
Takeuchi M, Seto T, Hashimoto M, Ichihara N, Morimoto Y, Kawakubo H, et al. Performance of a deep learning-based identification system for esophageal cancer from CT images. Esophagus. 2021;18:612–20.
Huang C, Dai Y, Chen Q, Chen H, Lin Y, Wu J, et al. Development and validation of a deep learning model to predict survival of patients with esophageal cancer. Front Oncol. 2022;12:971190.
Chen C, Ma Y, Zhu M, Yan Z, Lv X, Chen C, et al. A new method for Raman spectral analysis: decision fusion-based transfer learning model. J Raman Spectrosc. 2023;54(3):314–23.
Rezvy S, Zebin T, Braden B, Pang W, Taylor S, Gao XW. Transfer learning for Endoscopy disease detection and segmentation with mask-RCNN benchmark architecture. In: CEUR Workshop Proceedings. CEUR-WS; 2020. pp. 68–72.
Ling Q, Liu X. Image recognition of esophageal Cancer based on ResNet and transfer learning. Int Core J Eng. 2022;8(5):863–9.
Mahboubisarighieh A, Shahverdi H, Jafarpoor Nesheli S, Alipoor Kermani M, Niknam M, Torkashvand M, et al. Assessing the efficacy of 3D Dual-CycleGAN model for multi-contrast MRI synthesis. Egypt J Radiol Nucl Med. 2024;55(1):1–12.
Huang G, Zhu J, Li J, Wang Z, Cheng L, Liu L, et al. Channel-attention U-Net: channel attention mechanism for semantic segmentation of esophagus and esophageal cancer. IEEE Access. 2020;8:122798–810.
Cai Y, Wang Y. Ma-unet: An improved version of unet based on multi-scale and attention mechanism for medical image segmentation. In: Third international conference on electronics and communication; network and computer technology (ECNCT 2021). SPIE; 2022. pp. 205–11.
Mali SA, Ibrahim A, Woodruff HC, Andrearczyk V, Müller H, Primakov S, et al. Making radiomics more reproducible across scanner and imaging protocol variations: a review of harmonization methods. J Pers Med. 2021;11(9):842.
Pfaehler E, Zhovannik I, Wei L, Boellaard R, Dekker A, Monshouwer R, et al. A systematic review and quality of reporting checklist for repeatability and reproducibility of radiomic features. Phys Imaging Radiat Oncol. 2021;20:69–75.
Jha AK, Mithun S, Jaiswar V, Sherkhane UB, Purandare NC, Prabhash K, et al. Repeatability and reproducibility study of radiomic features on a Phantom and human cohort. Sci Rep. 2021;11(1):2055.
Xie C, Yu X, Tan N, Zhang J, Su W, Ni W, et al. Combined deep learning and radiomics in pretreatment radiation esophagitis prediction for patients with esophageal cancer underwent volumetric modulated Arc therapy. Radiother Oncol. 2024;199:110438.
Chen L, Ouyang Y, Liu S, Lin J, Chen C, Zheng C, et al. Radiomics analysis of lymph nodes with esophageal squamous cell carcinoma based on deep learning. J Oncol. 2022;2022(1):8534262.
Yang M, Hu P, Li M, Ding R, Wang Y, Pan S. Computed tomography-based Radiomics in Predicting T stage and length of esophageal squamous cell carcinoma. Front Oncol. 2021; 11: 722961. 2021.
Du KP, Huang WP, Liu SY, Chen YJ, Li LM, Liu XN, et al. Application of computed tomography-based radiomics in differential diagnosis of adenocarcinoma and squamous cell carcinoma at the esophagogastric junction. World J Gastroenterol. 2022;28(31):4363.
Acknowledgements
Authors are grateful to the Researchers Supporting Project (ANUI2024M111), Alnoor University, Mosul, Iraq.
Funding
None.
Author information
Authors and Affiliations
Contributions
M.A., H.H.A., R.A.K., S.G., A.Y., A.Sh., S.G., K.K.J., H.N.S., A.Y., Z.H.A. and A.M.: Investigation; Project administration; Methodology; Software; Formal analysis; Writing-original draft. B.F.: Conceptualization; Data curation; Project administration; Conceptualization; Supervision; Writing-review & editing. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethical approval
The need for ethical approval was waived off by the ethical committee of Alnoor University, Nineveh, Iraq. This study was conducted in accordance with the Declaration of Helsinki.
Consent to participate
It was waived off by the ethical committee of Alnoor University, Nineveh, Iraq.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Alsallal, M., Ahmed, H.H., Kareem, R.A. et al. A novel framework for esophageal cancer grading: combining CT imaging, radiomics, reproducibility, and deep learning insights. BMC Gastroenterol 25, 356 (2025). https://doiorg.publicaciones.saludcastillayleon.es/10.1186/s12876-025-03952-6
Received:
Accepted:
Published:
DOI: https://doiorg.publicaciones.saludcastillayleon.es/10.1186/s12876-025-03952-6